Report of the
High Data-Rate Macromolecular Crystallography Meeting
ACA 2019, Covingon, KY 21 July 2019
Report Date: 11 August 2019
This is a report of the informal High Data Rate Macromolecular Crystallography (HDRMX) dinner meeting during the ACA meeting in Covington, Kentucky
from 8 pm to 10:30 pm on 21 July 2019 at at Fire at RiverCenter in Covington, 50 E.
RiverCenter Blvd, Covington, KY.
There was an informal HDRMX dinner meeting during the ACA meeting in Covington, Kentucky from 8 pm to 10:30 pm on Sunday, 21 July 2019, at Fire at RiverCenter in Covington, 50 E.
RiverCenter Blvd, Covington, KY.
Herbert J. Bernstein, Ronin Institute
The changes are in two parts -- mandatory metadata such as full axis chain descriptions to be added
in a Dectris supported template that should appear in all MX data collected after
adoption of the gold standard so that data collected at any beamline following that standard would
be feasible to process using just what is in the data and metadata files without reference
to additional site files or lab notebooks, and optional additional metadata that individual
beamlines and users consider appropriate to add for other purposes, such sequence information
in preparation for map threading and future PDB deposition.
After vigorous discussion, it was agreed to use as the gold standard the NXmx-compliant metadata
exemplified by the latest DLS NeXus data files
from Graeme Winter, augmented by metadata exemplified by the latest
LCLS NeXus data files from Aaron Brewster, with the additional specifications
that the NXinstrument group would have the name of the beamline and the
NXsource group would have the name of the facility, and that NXlog would be used to provide accurate
timestamps for images. Graeme Winter and Aaron Brewster agreed to serve as a working
group to provide full example files
and to update the NXmx application definition with the details of these changes, so that gold
standard metadata files can be validated against the
augmented NXmx. It was agreed that the optional data would include any and all imgcif and/or PDB
mmcif/pdbx dictionary tags not already defined by NXmx using the recently defined NXpdb
group which, at the request of NIAC, will be validated against the relevant dictionaries, rather
than against NXmx.
Note that under an earlier agreement between NIAC and COMCIFS, and to allow CIF templates to be used
with NeXus files, we will conform the CBF/imgcif dictionary to any NeXus NXmx changes
with appropriate specific metadata tag mapping (e.g. the changes needed to use of the McStas coordinate
system in NeXus and the modified Mosflm coordinate system in CBF/imgcif). This way
it should be feasible to translate back and forth between Pilatus full-CBF and Eiger NeXus datasets as
needed.
It is hoped that progress will be sufficiently rapid to allow discussion of this standard at ECM32 and
adoption at a formal HDRMX meeting in early November 2019 at Diamond Light Source.
There are signs of divergence in Eiger formats among beamlines, and it is time to add new metadata, for
example to identify beamlines and facilities and to record metadata that will be
helpful in PDB depositions.
The primary objective is to ensure that sufficient metadata will be provided to allow processing at
a facility other than the one at which the data was produced. In particular, detailed
descriptions of axis chains to be used to process the data are needed, both for sample goniometers
and detector positioners.
In general, the requested augmentation of metadata is divided into two groups:
first, metadata to be added via a templating mechanism in the Dectris software to be set-up before
collection as static changes to the "master" files, and, second, metadata to be added
after collection, possibly via H5copy. For simplicity we refer to the former as static and the latter as
dynamic.
Some tags for static (i.e. Dectris template) additions are already available. imgCIF defines AXIS
tags needed for specification of arbitrary and very general axis chains. NeXus defines the
equivalent information in the NXtransformations base class. Concern has been expressed about
cluttering the templating mechanism with large numbers of tags used only in the most complex
cases.
To avoid such clutter, the input to the template can be the path to either a CBF or a NeXus
file with the appropriate axis information, along with the necessary software to automatically
convert between CBF and NeXus axis conventions. One way or another all diffraction geometry and
all detector geometry need to be described.
Tags have been defined to carry metadata specifying the beamline and facilty in CBF templates,
which will automatically map to the NeXus NXinstrument and NXsource name fields.
Note that the detector distance, wavelength and beam center are already specified and very necessary.
As integrating detectors or other detectors that do not count single photons come into use
in this performance range, detector gain will need to be specified.
Tags are needed for the HDF5 software version, to declare the use of non-standard local
format conventions, to list the files comprising a dataset, and to give the format of each
particular file.
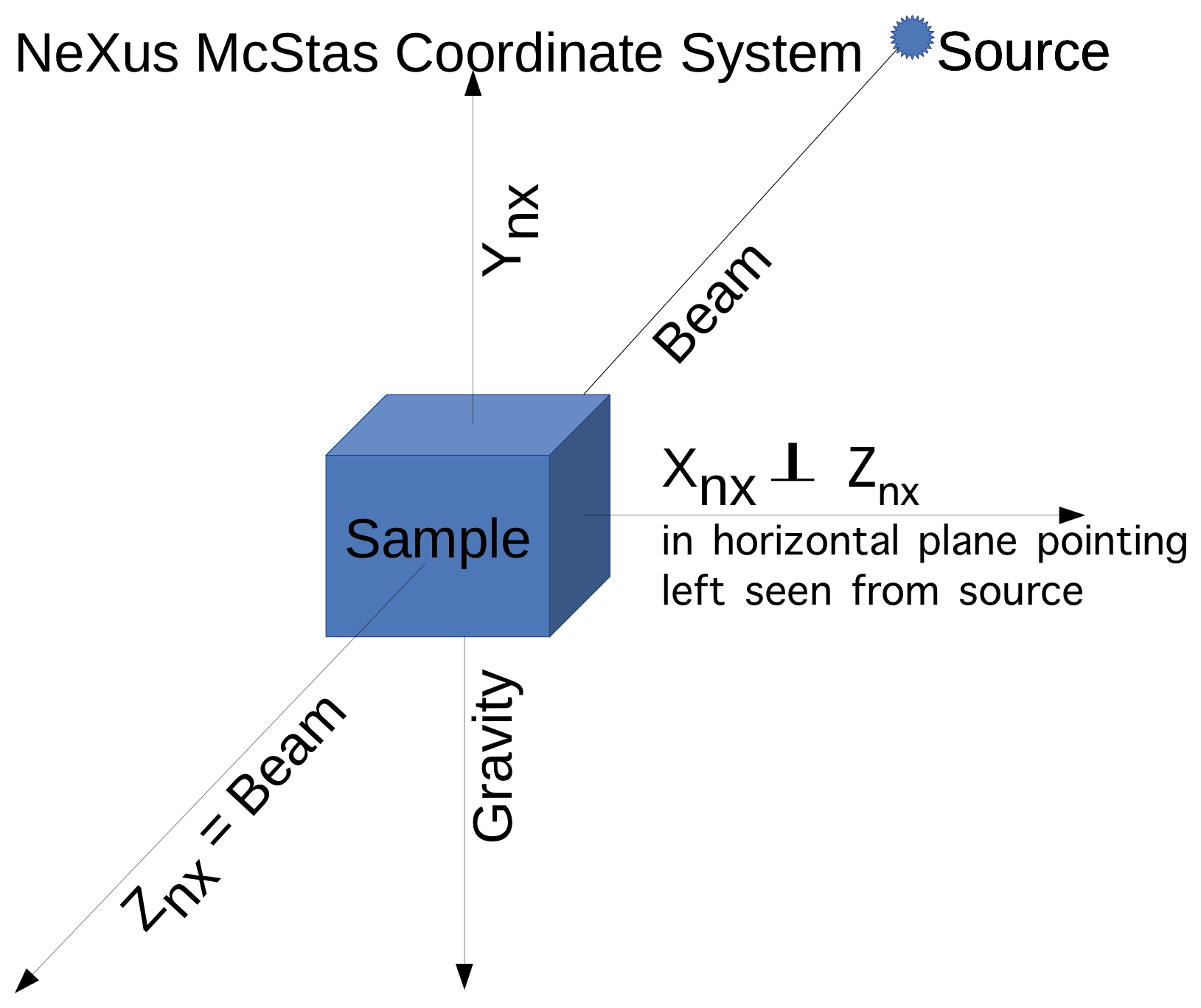
The NeXus/HDF5 files specify
axes in the NeXus McStas coordinate system.
The standard coordinate frame in NeXus is the McStas coordinate frame,
in which the Z axis points in the direction of the incident beam, the
X axis is orthogonal to the Z axis in the horizontal plane and pointing
left as seen from the source and the Y axis points upwards. The
origin is in the sample.
The standard coordinate frame in imgCIF/CBF aligns the X axis to the
principal goniometer axis, chooses the Z axis to point from the sample
into the beam.
Many tags for dynamic (non-Dectris-template) additions are also already available. For example, the monochrometer, the
beam_height, beam_width, beam_flux and sample sequence can all be
placed by a beamline or user in a CIF or NeXus file for merging with H5copy into an existing master metadata file. The
existing imgcif and mmcif dictionaries provide appropriate tags to
use, and more can be added.
Tags are already available to record frame exposure times, but some further discussion is needed
to specify what should be mandatory and what should be recommended best practice going beyond
the minimum required. In many cases it may be sufficient to record simple average
frame exposure times and periods, but as data collection rates continue to rise, it may be
necessary for some experiments at some facilities to record precise frame-by-frame times.
Aaron Brewster has posted a Jungfrau 16M dataset as an example dataset here:
https://doi.org/10.5281/zenodo.3352357
The HDRMX website is http://hdrmx.medsbio.org. There will also
be an informal HDRMX
meeting at Zum Leupold in Vienna at 19:30 on 20 August 2019. Contact H. J. Bernstein at
yayahjb at gmail dot com if you wish to participate to see if there is still
space.
Participants:
Lawrence C. Andrews, Ronin Institute
Frances C. Bernstein, Bernstein+Sons
Aaron Brewster, LBNL
Andreas Förster, Dectris
Ana Gonzalez, MaxIV
Pascal Hofer, Dectris
James Holton, ALS (partial attendance)
Loes Kroon-Batenburg, Utrecht University
Filip Leonarski, PSI
Art Lyubimov, SLAC
Katherine McAuley, DLS
Clemens Vonrhein, Global Phasing
Graeme Winter, DLS
Discussion
The main topic for discussion was the pending changes to the NeXus/HDF5 format to create a new
"gold standard" as well as the interaction with Eiger2.
General Sense of the Covington Meeting
Structure of the New Metadata
Static Metadata
Main Points of the Agreement
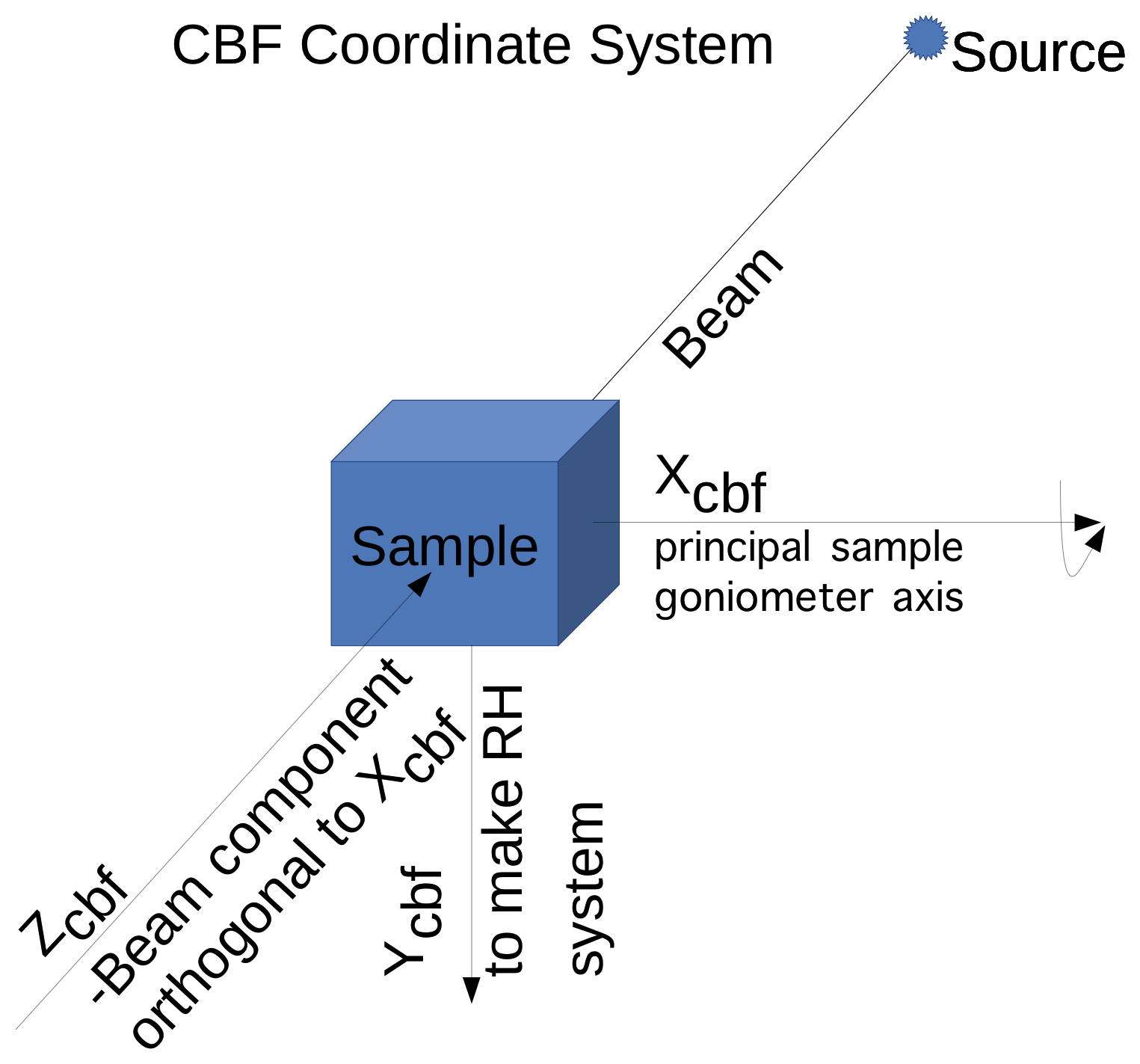 If the beam is not orthogonal to the X axis, the Z axis
is the component orthogonal to the X axis the of "-Beam" vector. The "-Beam"
vector
is the negative of the "Beam" vector, i.e. a vector which points towards
the source. The Y axis is chosen to complete a right-handed axis system.
If the beam is not orthogonal to the X axis, the Z axis
is the component orthogonal to the X axis the of "-Beam" vector. The "-Beam"
vector
is the negative of the "Beam" vector, i.e. a vector which points towards
the source. The Y axis is chosen to complete a right-handed axis system.
Dynamic Metadata
sample provenance, sample physical characteristics, sample imagery, protein sequence, detector
and sample environments, incl. temperature, sample delivery method, serial
crystallography parameters (incl. pump probes), spectroscopy, sample mount, detector ROI.
beamline optics, source parameters, e.g. mode, current, collection strategy, scan type, scan mode,
beam profile (Gaussian, tophat), monochromator bandpass, beam divergences, and beam collimation.
Partial Example
As a partial example consider a beamline called XXX (ID1) at site SYNC with an omega axis, and pin_x, pin_y and pin_z
translation axes stacked 5 millimetres apart, using hdf5_1.8.14 and
NXmx 1.4. Then a portion of the necessary information presented as a CIF file might be:
data_AMX_metadata
loop_
_axis.id
_axis.type
_axis.equipment
_axis.depends_on
_axis.vector[1]
_axis.vector[2]
_axis.vector[3]
_axis.offset[1]
_axis.offset[2]
_axis.offset[3]
source . source . 0 0 1 . . .
gravity . gravity . 0 -1 0 . . .
pin_x translation goniometer . -1 0 0 0 0 0
omega rotation goniometer pin_x 1 0 0 -5 0 0
pin_y rotation goniometer omega 0 1 0 -10 0 0
pin_z rotation goniometer pin_y 0 0 -1 -15 0 0
_array_intensities.gain 1.0 #counts/photon
_diffrn_source.source SYNCHROTRON
_diffrn_source.type 'SYNC XXX (ID1)'
_diffrn_source.pdbx_synchrotron SYNC
# to be mapped to NXsource/name
_diffrn_source.pdbx_synchrotron_beamline 'XXX (ID1)'
# to be mapped to NXinstrument/name
_dataset_file_format.file_format 'hdf5_1.8.14 and NXmx 1.4'
_diffrn_radiation.beam_width 7 #micrometres
_diffrn_radiation.beam_height 5 #micrometres
_diffrn_radiation.beam_flux 400000000000 #ph/s in the beam
Time -- an issue to be resolved
Sample Data
How to Get Involved